Users download software on as many as 700,000 home PCs to help crack the code on the coronavirus.
by RICK MERRITT
The tweet party started on Wednesday, March 25, at 1:55pm.
That’s when the research network that Folding@Home manages had arguably become the world’s most powerful supercomputer. Its call to help fight the coronavirus on March 15 had amassed enough donations of spare cycles on home computers to create a crowd-sourced exascale supercomputer.
In just 10 days, supporters had downloaded the group’s software on hundreds of thousands of home PCs to help crack the code on COVID-19.
“It’s been a pretty amazing experience,” said Greg Bowman, director of Folding@Home, an all-volunteer team of researchers. Bowman’s also an associate professor at the Washington University School of Medicine, in St. Louis, home to one of 11 labs worldwide that keep the Folding@Home network humming.
“It’s exciting to have the chance to try everything we can think of in parallel to maximize the chances of having an impact on the current pandemic,” he said.
Supporters sheltering at home posted a virtual party of animations in response to the tweet that announced the news. Schoolkids in video clips danced on their desks, Top Gun heroes slapped high-fives, the Cookie Monster gobbled goodies, and a small army of Star Trek, Star Wars and anime characters whooped it up. Even an animated Simon Cowell offered a thumbs up.
At Folding@Home, the leaderboard continues to rise. More than a million computers are now on the network that started the year with just 30,000.
Bowman estimates performance has surpassed 1.5 exaflops, fueled in part by more than 356,000 NVIDIA GPUs. The group’s blog site will share more news soon, he said.
Sneak Peek of a COVID-19 Spike
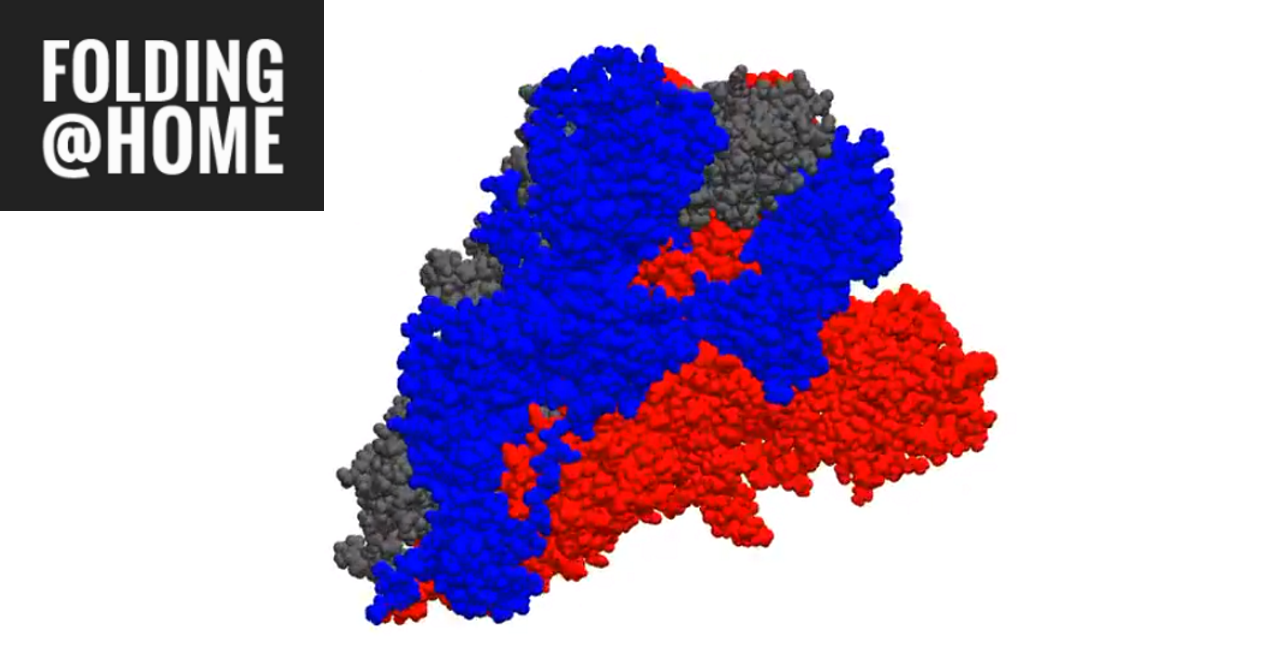
Even before it broke the exaflop barrier, researchers were posting eye-popping work. An animation of a coronavirus protein that infects healthy cells could suggest molecular targets for an anti-COVID drug or vaccine (see the image above).
The effort builds on the group’s earlier experiments on the Ebola virus. It’s part of its broad work exploring how proteins – a basic ingredient of living things including viruses – fold into 3D shapes to perform various functions.
The first snapshot of a coronavirus protein couldn’t be captured with a simple smartphone selfie. Such scientific visualizations demand massive compute power to simulate near-atomic-level interactions.
Flipping on an Exaflop
These days, computer scientists stack computer rooms the size of a couple tennis courts with servers to gnaw on big questions of science. The Summit system at Oak Ridge National Laboratory, currently the world’s largest supercomputer, packs 27,648 NVIDIA Tensor Core GPUs to deliver 148.6 petaflops on the HPL benchmark. The second-ranked Sierra system at Lawrence Livermore National Laboratory also uses NVIDIA GPUs to crank out 94.6 petaflops.
The U.S. Department of Energy, which runs both systems, aims to switch on its first exascale system sometime in 2021. Arguably, the exascale era has already begun. Summit has racked up a handful milestones, including a 3.3 exaflops score running mixed-precision AI tasks.
That said, Folding@Home gets kudos as the first crowd-sourced effort to assemble exascale horsepower.
It’s a heady achievement. Imagine a billion people, each holding a billion calculators. If they all hit the equal sign at the same time, they’d execute in one second a quintillion calculations. That’s one exaflop.
Many Ways You Can Help
Trying to ride the tsunami of donations, Bowman told supporters over Twitter that researchers briefly scrambled for more servers to keep all the connected PCs fed.
“I hope I can speak for most people when I say this has given me something that I can contribute and has had a positive effect on my mental state,” replied one backer.
“Folding has on and off heated the top floor of my house,” quipped a computer specialist in the U.K.
The group continues to encourage users to donate time on their PCs with software they can download here. In turn, Folding@Home makes data from its experiments publicly available on open general research sites and ones devoted to areas such as biology.
To accelerate its efforts, Folding@Home recently announced a new batch of small molecule screening simulations that will help prioritize which molecules will be synthesized and assayed by the COVID Moonshot.
Folding@Home is one of many technology efforts formed to fight coronavirus.
The U.S. Department of Energy created the COVID-19 High Performance Computing Consortium with support from many tech companies and universities. The group will pool 16 supercomputers to deliver a combined 330 petaflops in part with help from more than 36,000 NVIDIA GPUs.
Separately, developers are already coding up projects in the #BuildforCOVID19 hackathon. Highlighted projects will be announced April 10.
For its part, NVIDIA is offering researchers a free 90-day license to its Parabricks software to accelerate the analysis of the coronavirus’ genetic code.
Users download software on as many as 700,000 home PCs to help crack the code on the coronavirus.
by RICK MERRITT

The tweet party started on Wednesday, March 25, at 1:55pm.
That’s when the research network that Folding@Home manages had arguably become the world’s most powerful supercomputer. Its call to help fight the coronavirus on March 15 had amassed enough donations of spare cycles on home computers to create a crowd-sourced exascale supercomputer.
In just 10 days, supporters had downloaded the group’s software on hundreds of thousands of home PCs to help crack the code on COVID-19.
“It’s been a pretty amazing experience,” said Greg Bowman, director of Folding@Home, an all-volunteer team of researchers. Bowman’s also an associate professor at the Washington University School of Medicine, in St. Louis, home to one of 11 labs worldwide that keep the Folding@Home network humming.
“It’s exciting to have the chance to try everything we can think of in parallel to maximize the chances of having an impact on the current pandemic,” he said.
Supporters sheltering at home posted a virtual party of animations in response to the tweet that announced the news. Schoolkids in video clips danced on their desks, Top Gun heroes slapped high-fives, the Cookie Monster gobbled goodies, and a small army of Star Trek, Star Wars and anime characters whooped it up. Even an animated Simon Cowell offered a thumbs up.
At Folding@Home, the leaderboard continues to rise. More than a million computers are now on the network that started the year with just 30,000.
Bowman estimates performance has surpassed 1.5 exaflops, fueled in part by more than 356,000 NVIDIA GPUs. The group’s blog site will share more news soon, he said.
Sneak Peek of a COVID-19 Spike
Even before it broke the exaflop barrier, researchers were posting eye-popping work. An animation of a coronavirus protein that infects healthy cells could suggest molecular targets for an anti-COVID drug or vaccine (see the image above).
The effort builds on the group’s earlier experiments on the Ebola virus. It’s part of its broad work exploring how proteins – a basic ingredient of living things including viruses – fold into 3D shapes to perform various functions.
The first snapshot of a coronavirus protein couldn’t be captured with a simple smartphone selfie. Such scientific visualizations demand massive compute power to simulate near-atomic-level interactions.
Flipping on an Exaflop
These days, computer scientists stack computer rooms the size of a couple tennis courts with servers to gnaw on big questions of science. The Summit system at Oak Ridge National Laboratory, currently the world’s largest supercomputer, packs 27,648 NVIDIA Tensor Core GPUs to deliver 148.6 petaflops on the HPL benchmark. The second-ranked Sierra system at Lawrence Livermore National Laboratory also uses NVIDIA GPUs to crank out 94.6 petaflops.
The U.S. Department of Energy, which runs both systems, aims to switch on its first exascale system sometime in 2021. Arguably, the exascale era has already begun. Summit has racked up a handful milestones, including a 3.3 exaflops score running mixed-precision AI tasks.
That said, Folding@Home gets kudos as the first crowd-sourced effort to assemble exascale horsepower.
It’s a heady achievement. Imagine a billion people, each holding a billion calculators. If they all hit the equal sign at the same time, they’d execute in one second a quintillion calculations. That’s one exaflop.
Many Ways You Can Help
Trying to ride the tsunami of donations, Bowman told supporters over Twitter that researchers briefly scrambled for more servers to keep all the connected PCs fed.
“I hope I can speak for most people when I say this has given me something that I can contribute and has had a positive effect on my mental state,” replied one backer.
“Folding has on and off heated the top floor of my house,” quipped a computer specialist in the U.K.
The group continues to encourage users to donate time on their PCs with software they can download here. In turn, Folding@Home makes data from its experiments publicly available on open general research sites and ones devoted to areas such as biology.
To accelerate its efforts, Folding@Home recently announced a new batch of small molecule screening simulations that will help prioritize which molecules will be synthesized and assayed by the COVID Moonshot.
Folding@Home is one of many technology efforts formed to fight coronavirus.
The U.S. Department of Energy created the COVID-19 High Performance Computing Consortium with support from many tech companies and universities. The group will pool 16 supercomputers to deliver a combined 330 petaflops in part with help from more than 36,000 NVIDIA GPUs.
Separately, developers are already coding up projects in the #BuildforCOVID19 hackathon. Highlighted projects will be announced April 10.
For its part, NVIDIA is offering researchers a free 90-day license to its Parabricks software to accelerate the analysis of the coronavirus’ genetic code.